My Apollo window has frozen. What do I do?
Try reloading your browser window.
- For Chrome: Shift Command R
- For Firefox: Ctrl+R
- For Safari: shift+reload
Some data tracks are very slow to load.
The way JBrowse works within Apollo, the first time your load up a track it might take a while. But once loaded, any successive loads will be quicker.
I used to see tracks and now they are gone.
Close the track and open it again. Yes, this is the standard 'turn it off and turn it on again' solution.
What is the meaning of the lower case letters in the nucleotide sequence?
These indicate regions of low complexity. They have no impact/implication with respect to annotation.
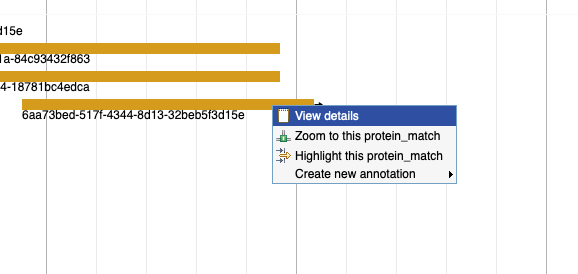
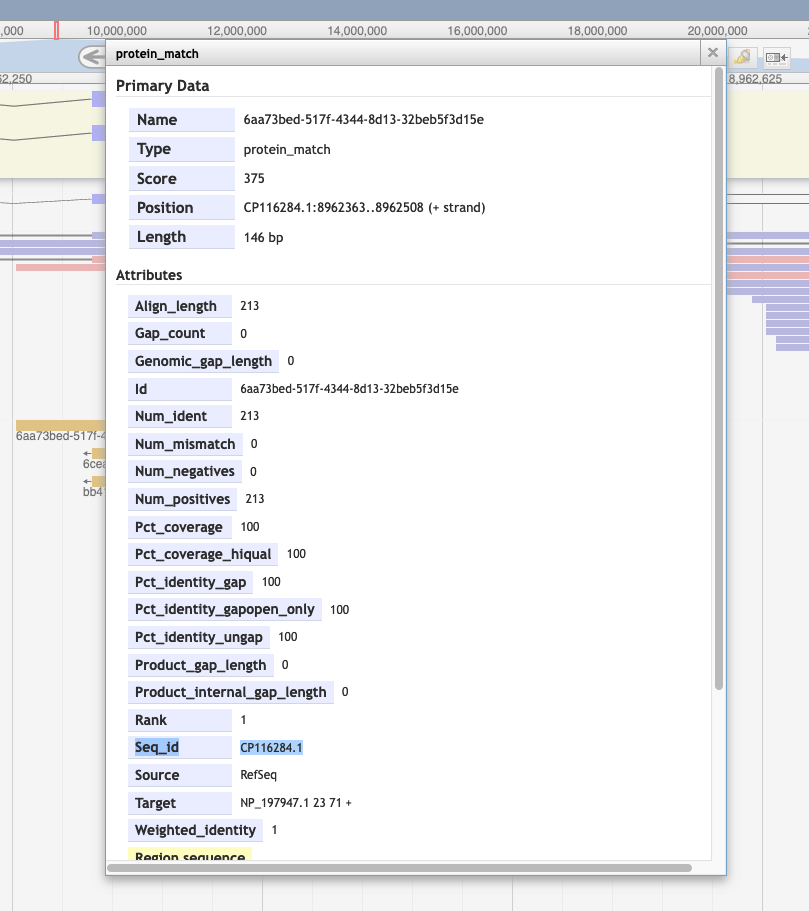
Is it possible to know which organism is the source for a particular protein alignment (in the Protein Alignment track)?
By right clicking on the alignment, and then selecting 'View details.' You'll see the GenBank accession in the id section and can search for that at NCBI.
Tell me more about the tracks I see.
Track name | Description of data |
Col-CC_Genomic_Annotations_Data | Result of NCBI Eukaryotic Annotation Pipeline |
Gnomon Models | One of the inputs into the NCBI Pipeline. "Gnomon annotation of the genomic sequence. Sequence identifiers are provided as accession.version for the genomic sequences and Gnomon identifiers for the Gnomon models:gene.XXX for genes, GNOMON.XXX.m for transcripts and GNOMON.XXX.p for proteins. These identifiers are NOT universally unique. They are unique per annotation release only." (from NCBI documentation) |
| Protein alignment | proteins from other species aligned to v12 protein sequences. "Alignments of cDNAs, ESTs and TSAs from other species to the genomic sequence(s). These alignments may have been used as evidence for gene prediction by the annotation pipeline." (from NCBI documentation) |
| TSA alignment | computationally assembled transcripts from isoseq experiments aligned to v12 transcript sequences |
AT-Col-CC-Liftoff-from-TAIR10.1 | v11 models mapped to v12 reference using Liftoff |
| Col-CC Same Species | "Alignments of same-species cDNAs, ESTs and TSAs to the genomic sequence(s). These alignments were used as evidence for gene prediction by the annotation pipeline. Sequence identifiers are provided as accession.version." (from NCBI documentation) |
| Known Reference Sequences | "Alignments of the annotated Known RefSeq transcripts (identified with accessions prefixed with NM_ and NR_) to the genome." (from NCBI documentation) |
| Model Reference Sequences | "Alignments of the annotated Model RefSeq transcripts (identified with accessions prefixed with XM_ and XR_) to the genome." (from NCBI documentation) |
| (RNA seq tracks) | (coming) |
| (long read seq tracks) | (coming) |
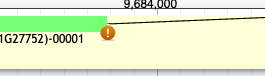
What does the warning symbol mean?
This symbol means that the location and sequence of the splice site needs to be investigated and verified because it does not conform to the GT-AG rule.
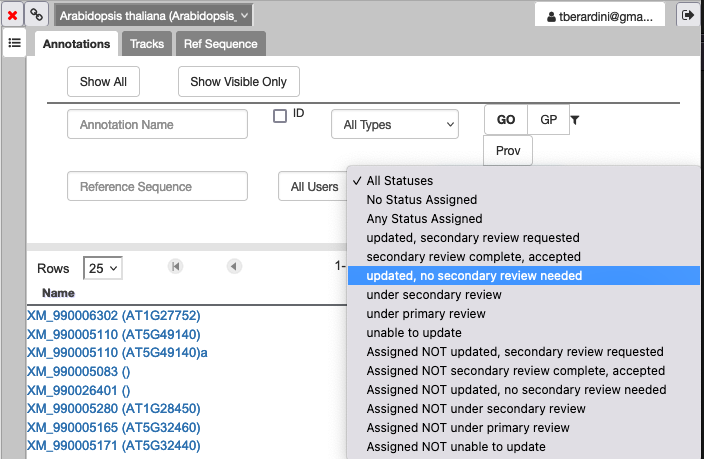
How do I see genes that need secondary review?
In the Annotations tab of your right hand panel, click on the dropdown for "All Statuses" and select the status you want to filter the annotations to review.
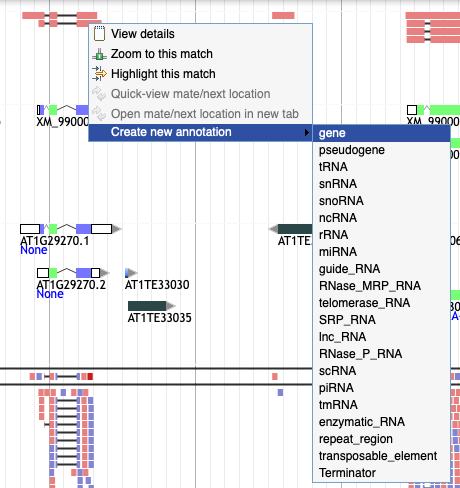
I want to drag an element from an evidence track to the user-created annotation track but I can't!
Right click on the element and select 'Create new annotation' from the menu. Pick the type of element you want to create and it will appear in the yellow track.